In bio-organic chemistry, a hexose is a monosaccharide with six carbonatoms, having the chemical formula C6H12O6. Hexoses are classified by functional group, with aldohexoses having an aldehyde at position 1, and ketohexoses having a ketone at position 2.
Aldohexoses
Ketohexoses
Mutations
The aldehyde and ketone functional groups in these carbohydrates react with neighbouring hydroxyl functional groups to form intramolecular hemiacetals and hemiketals, respectively. The resulting ring structure is related to pyran, and is termed a pyranose. The ring spontaneously opens and closes, allowing rotation to occur about the bond between the carbonyl group and the neighbouring carbon atom, yielding two distinct configurations (α and β). This process is termed mutarotation. Hexose sugars can form dihexose sugars with a condensation reaction to form a 1,6-glycosidic bond.
Protein-hexose binding
References
Wikipedia
Aldohexoses
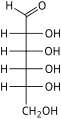
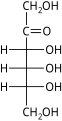
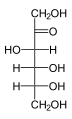
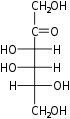
The aldohexoses have four chiral centres for a total of 16 possible aldohexose stereoisomers (24). The D/L configuration is based on the orientation of the hydroxyl at position 5, and does not refer to the direction of optical activity. The eight D-aldohexoses are:
Of these D-isomers, all except D-altrose are naturally occurring. L-Altrose, however, has been isolated from strains of the bacterium Butyrivibrio fibrisolvens.[1]
A mnemonic for the aldohexoses is "All Altruists Gladly Make Gum in Gallon Tanks": allose, altrose, glucose, mannose, gulose, idose, galactose, talose. When drawn in this order, the Fischer projections of the D-aldohexoses follow a pattern: beginning with allose having all four hydroxyl groups on the right, going through the mnemonic at carbon 2 the hydroxyl groups alternate right-left with each different sugar (allose left, altrose right, glucose left, etc.). At carbon 3, the first two are on the right, the next two are on the left, and so on. At carbon 4, the first four are on the right and the rest are on the left. At carbon 5, all eight D-aldohexoses have the hydroxyl group on the right (because if the hydroxyl group were on the left instead, due to the d/l system naming convention, they would be L-enantiomers of the D versions seen above).
This can be seen as binary counting to seven, where 0 stands for hydroxyl and 1 for hydrogen. So 0000 stands for D-Allose, 0001 stands for D-Altrose, 0010 stands for D-Glucose, 0011 stands for D-Mannose, 0100 stands for D-Gulose, 0101 stands for D-Idose, 0110 stands for D-Galactose and 0111 stands for D-Talose.
Cyclic hemiacetals
It has been known since 1926 that 6-carbon aldose sugars form cyclic hemiacetals.[2] The diagram below shows the hemiacetal forms for D-glucose and D-mannose.
The numbered carbons in the open-chain forms correspond to the same numbered carbons in the hemiacetal forms. The formation of the hemiacetal causes carbon number 1, which is symmetric in the open-chain form, to become asymmetric in the cyclic version. This means that both glucose and mannose (as well as all the other aldohexoses) each have two cyclic forms. In solution, both of these exist in equilibrium with the open-chain form. The open-chain form, however, does not crystallize. Hence the two cyclic forms become separable when they are crystallized. For example, D-glucose forms an alpha crystal that has specific rotation of +112° and melting point of 146 °C, as well as a beta crystal that has specific rotation of +19° and melting point of 150 °C.[2]
Ketohexoses
The ketohexoses have 3 chiral centres and therefore eight possible stereoisomers (23). Of these, only the four D-isomers are known to occur naturally:
Only the naturally occurring hexoses are capable of being fermented by yeasts.
Mutations
The aldehyde and ketone functional groups in these carbohydrates react with neighbouring hydroxyl functional groups to form intramolecular hemiacetals and hemiketals, respectively. The resulting ring structure is related to pyran, and is termed a pyranose. The ring spontaneously opens and closes, allowing rotation to occur about the bond between the carbonyl group and the neighbouring carbon atom, yielding two distinct configurations (α and β). This process is termed mutarotation. Hexose sugars can form dihexose sugars with a condensation reaction to form a 1,6-glycosidic bond.
Protein-hexose binding
Hexoses play a key role in several different biochemical pathways, including cellular energy release, signaling, carbohydrate synthesis, and the regulation of gene expression. The most common hexoses in biological systems are D-galactose, D-glucose and D-mannose.
Researchers have used computational algorithms to model protein-galactose,[3] protein-glucose[4] and protein-mannose[5] binding-sites. Individual hexoses share multiple physio-chemical similarities leading to the development of a general protein-hexose binding-site model.[6]
Using Inductive logic programming, a machine learning approach that learns if-then rules from data, Nassif et al. recovered known wet-lab hexose-binding spatial, physical and chemical properties from the Protein Data Bank.[7]
References
- ^ US patent 4966845, Stack; Robert J., "Microbial production of L-altrose", issued 1990-10-30, assigned to Government of the United States of America, Secretary of Agriculture
- ^ a b Morrison, Robert Thornton; Boyd, Robert Neilson. Organic Chemistry (2nd ed.). Allyn and Bacon. Library of Congress catalog 66-25695
- ^ Sujatha, MS; Balaji, PV (2004). "Identification of common structural features of binding sites in galactose-specific proteins.". Proteins: Structure, Function, and Bioinformatics 55(1): 44–65. doi:10.1002/prot.10612.
- ^ Nassif, Houssam; Al-Ali, Hassan; Khuri, Sawsan; Keyrouz, Walid (2009). "Prediction of Protein-Glucose Binding Sites Using Support Vector Machines". Proteins: Structure, Function and Bioinformatics 77 (7): 121–132. doi:10.1002/prot.22424. PMID 19415755.
- ^ Agarwal, Sandhya; Mishra, Nitish Kumar; Singh, Harinder; Raghava, Gajendra PS (2011). "Identification of Mannose Interacting Residues Using Local Composition". PLOS ONE 6 (9). doi:10.1371/journal.pone.0024039.
- ^ Santos, Jose; Nassif, Houssam; Page, David; Muggleton, Stephen; Sternberg, Mike (2012). "Automated identification of features of protein-ligand interactions using Inductive Logic Programming: a hexose binding case study" (PDF). BMC Bioinformatics 13: 162. doi:10.1186/1471-2105-13-162.
- ^ Nassif, Houssam; Al-Ali, Hassan; Khuri, Sawsan; Keyrouz, Walid; Page, David (2009). "An Inductive Logic Programming Approach to Validate Hexose Binding Biochemical Knowledge" (PDF). International Conference on Inductive Logic Programming (ILP'09): 149–165.
Wikipedia














No comments:
Post a Comment