Published Date
Author
For further details log on website :
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0152400
- Published: April 7, 2016
- http://dx.doi.org/10.1371/journal.pone.0152400
Author
Abstract
Cryptocercus punctulatus and Parasphaeria boleiriana are two distantly related xylophagous and subsocial cockroaches. Cryptocercus is related to termites. Xylophagous cockroaches and termites are excellent model organisms for studying the symbiotic relationship between the insect and their microbiota. In this study, high-throughput 454 pyrosequencing of 16S rRNA was used to investigate the diversity of metagenomic gut communities of C. punctulatus and P. boleiriana, and thereby to identify possible shifts in symbiont allegiances during cockroaches evolution. Our results revealed that the hindgut prokaryotic communities of both xylophagous cockroaches are dominated by members of four Bacteria phyla: Bacteroidetes, Firmicutes, Proteobacteria, and Actinobacteria. Other identified phyla were Spirochaetes, Planctomycetes, candidatus Saccharibacteria (formerly TM7), and Acidobacteria, each of which represented 1–2% of the total population detected. Community similarity based on phylogenetic relatedness by unweighted UniFrac analyses indicated that the composition of the bacterial community in the two species was significantly different (P < 0.05). Phylogenetic analysis based on the characterized clusters of Bacteroidetes, Spirochaetes, and Deltaproteobacteria showed that many OTUs present in both cockroach species clustered with sequences previously described in termites and other cockroaches, but not with those from other animals or environments. These results suggest that, during their evolution, those cockroaches conserved several bacterial communities from the microbiota of a common ancestor. The ecological stability of those microbial communities may imply the important functional role for the survival of the host of providing nutrients in appropriate quantities and balance.
Figures
Citation: Berlanga M, Llorens C, Comas J, Guerrero R (2016) Gut Bacterial Community of the Xylophagous Cockroaches Cryptocercus punctulatus and Parasphaeria boleiriana. PLoS ONE 11(4): e0152400. doi:10.1371/journal.pone.0152400
Editor: Lorenzo Brusetti, Free University of Bozen/Bolzano, ITALY
Received: May 25, 2015; Accepted: March 14, 2016; Published: April 7, 2016
Copyright: © 2016 Berlanga et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Data Availability: Data are available from NCBI at accession numbers: SRR2035371 and SRR2035362.
Funding: This study was funded by CGL2009-08922 (Spanish Ministry of Science and Technology) to Ricardo Guerrero. Author Carlos Llorens is employed by Biotechvana during the study. Biotechvana provided support in the form of salary for author CL, but did not provide specific funding for this study and did not have any additional role in the study design, data collection, decision to publish, or preparation of the manuscript. This specific role of this author is articulated in the ‘author contributions’ section.
Competing interests: Author Carlos Llorens is employed by Biotechvana. There are no patents, products in development or marketed products to declare. This does not alter the authors' adherence to all the PLOS ONE policies on sharing data and materials.
Introduction
Insects account for most of the richness of species of the animal clades on Earth. The associations between microorganisms and insects are widespread in nature [1]. For their insect hosts, bacteria can provide numerous benefits, such as specific nutritional complementation of a markedly imbalanced diet, protection from predators, parasites, and pathogens; and the promotion of mating and reproduction [2,3]. Termites (Isoptera), cockroaches, and mantids form a well-established lineage of insects, the Dictyoptera. In fact, termites are actually social cockroaches [4], being the family Cryptocercidae their closest relative and the Mantodea(mantids) the sister group to the clade comprising cockroaches and termites [5]. Six families of termites (collectively called lower termites) share with Cryptocercus spp. the unusual ability to degrade lignocellulosic plant material, carried out by the metabolic activities of the bacteria and protists of their gut microbiota [6–9]. Higher termites have lost their gut protists, having only bacteria. They are represented by a single highly diversified family, the Termitidae.
Modern cockroaches are thought to have radiated at some time between the late Jurassic and early Cretaceous, ~140 million years ago [10]. There are fundamental differences in the diets of termites and cockroaches. While termites feed almost exclusively on lignocellulose in various stages of decay, many cockroaches subsist on a highly variable diet. Examples of xylophagy in cockroaches are Cryptocercus spp. (family Cryptocercidae) from East Asia and North America, Panesthia spp. (subfamily Panesthiinae, family Blaberidae) in Australia and Asia, and Parasphaeria boleiriana (family Blaberidae) from Brazil [11].
In this study, we compared the bacterial gut microbiota of two xylophagous cockroaches, Cryptocercus puctulatus and Parasphaeria boleiriana. Members of the genus Cryptocercus are subsocial cockroaches that inhabit temperate forests of the northern hemisphere, living in extensive galleries excavated within decomposing logs. At present, nine species in the genus are recognized worldwide: two in eastern Eurasia, two in southwestern China, and five in the USA. The distribution of C. punctulatus extends throughout western Virginia and Pennsylvania. The ecological niche for the five Nearctic Cryptocercus species lies within a small range of the spectrum of annual mean temperatures and precipitation that characterize this region: 6–17°C and 140–470 mm/m2, respectively [12].
Parasphaeria boleiriana lives in the remnants of the semi-deciduous table land of the Atlantic forest in the state of Espirito Santo, Brazil. The genus was previously known for the species P. ovata, from Chile and Argentina. P. boleiriana feeds on the softwood of boleira (the tree Joannessia princeps). It differs from Cryptocercus in that it develops and reproduces in a very short time, 2–3 years, rather than >5 years, and survives as an adult for only one season, rather than several years. Brood care by P. boleiriana is also very short, with a mean of 12 days, compared to several years by Cryptocercus [11,13,14].
In lower termites and Cryptocercus cockroaches, wood is efficiently digested by their flagellate symbionts (eukaryotes), whereas in higher termites lignocellulosic material (wood, detritus, humus, etc.) is digested by a diverse assemblage of cellulolytic prokaryotes. The clade Cryptocercus–termites clearly shows the coevolution of the host with a stable intestinal microbiota essential to its survival [15–17]. By contrast, detailed information on the gut bacterial diversity in Parasphaeria (which is phylogenetically distinct from the clade Cryptocercus–termites) that allows lignocellulose digestion is still lacking. In the present work, the diversity of metagenomic gut communities of C. punctulatus and P. boleiriana was investigated to identify possible shifts in symbiont allegiances during cockroach evolution. We also compared several bacterial phyla associated with the microbiota of cockroaches with other metagenomes databases that correspond on Cryptocercus, termites and other insect groups.
Materials and Methods
Cockroaches and isolation of bacterial DNA
Cryptocercus punctulatus was collected from Virginia (USA) by Dr. Michael Dolan (University of Massachusetts at Amherst, MA, USA), and Parasphaeria boleiriana from Brazil by Dr. Philippe Grandcolas (CNRS-National Museum of Natural History, Paris, France). Two individuals of each species were sent to our laboratory in Barcelona, Spain. During transport, the cockroaches were maintained in tubes with wood at room temperature. Immediately after their arrival in the laboratory, they were dissected to extract the whole gut. Cockroaches were dissected with a sterile scalpel. The abdomens of the insects were incised to remove the dorsal cuticle, the gut was collected under sterile de-ionized water, and the hindgut region was separated and placed into an Eppendorf tube for DNA extraction. The hindgut of the insect was homogenized using a FastPrep system (MP Biomedicals Europe) with 0.1-mm glass beads. Bulk DNA was extracted by several washings with phenol-chloroform [18]. All material and solutions used were sterile. Disinfection and dissection were performed in a laminar flow cabinet. During extraction, we worked in an aseptic environment under laminar hood to avoid contamination [19].
Amplicon library preparation
Amplification of the variable region V1–V2 of the bacterial 16S rDNA gene was utilized to assess gut microbial diversity. Primers used were 8F-338R (5′-AGAGTTTGATCCTGGCTCAG-3′ and 5′-TGCTGCCTCCCGTAGGAGT-3′) for multiplex Roche 454 GS FLX pyrosequencing. Primer design was carried out according to the manufacturer’s instructions. Initial PCR from each DNA was performed four times [20,21]. After PCR, the resulting product was checked for size and purity on an agarose-Sybr safe DNA gel stain (Invitrogen, San Diego, CA, USA). The amplicons were purified using a Pure Link kit (Invitrogen, San Diego, CA, USA) and quantified using Qubit and Bioanalyzer. The pool of amplicons were mixed equimolar (four amplicons for cockroach specie) and then prepared for 454-pyrosequencing according to the manufacturer. Cycling conditions were 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 56°C for 40 s, 68°C for 40 s, and a final extension step at 68°C for 6 min.
Bioinformatic analyses
Raw data of both cockroach metagenomes obtained were 5188 and 5788, Cryptocercus and Parasphaeria, respectively. Data were preprocessed for demultiplex and quality control using a pipeline implemented in GPRO version 1.1 [22]. This pipeline combines the tools Cutadapt [23], Prinseq-Lite [24], and FastQC [25]. Reads less than 250 nucleotides in size and redundant sequences were removed from each metagenome dataset using GPRO and Mothur1.31.2 [26]. This approach resulted in a non-redundant database of 3519 sequences from the Cryptocercusdataset and 2744 sequences from the Parasphaeria dataset. Data deposition: Bioproject PRJNA284583.
A multiple alignment was constructed for each dataset using the secondary-structure aware infernal aligner [27] combined with Genedoc [28] for manual refinement. Sequences not fulfilling at least 80% of the common core and gaps and non-informative traits were filtered from each alignment by combining the “unique.seqs,” “screen.seqs,” and “filter.seqs” commands of Mothur. CD-HIT-EST from the CD-HIT 4.5.4 package [29] was subsequently used to define clusters of clones within each metagenome with a distance threshold of 0.03 (resulting in a cut-off at the species level). The 3.69 Phylip Dnadist tool [http://evolution.gs.washington.edu/phylip.html] was used to obtain the neighbor-joining (NJ) distance matrix for each alignment. Both matrices were subsequently used to obtain rarefaction curves at different distances (0.03, 0.05, 0.10, and 0.15) and several diversity indices using Mothur1.36.1. Taxonomy was assigned by the Silva database [http://www.arb-silva.de] [30]. Community comparison of both metagenome Cryptocercus and Parasphaeria was evaluated using the UniFrac Server [31].
Statistical analyses
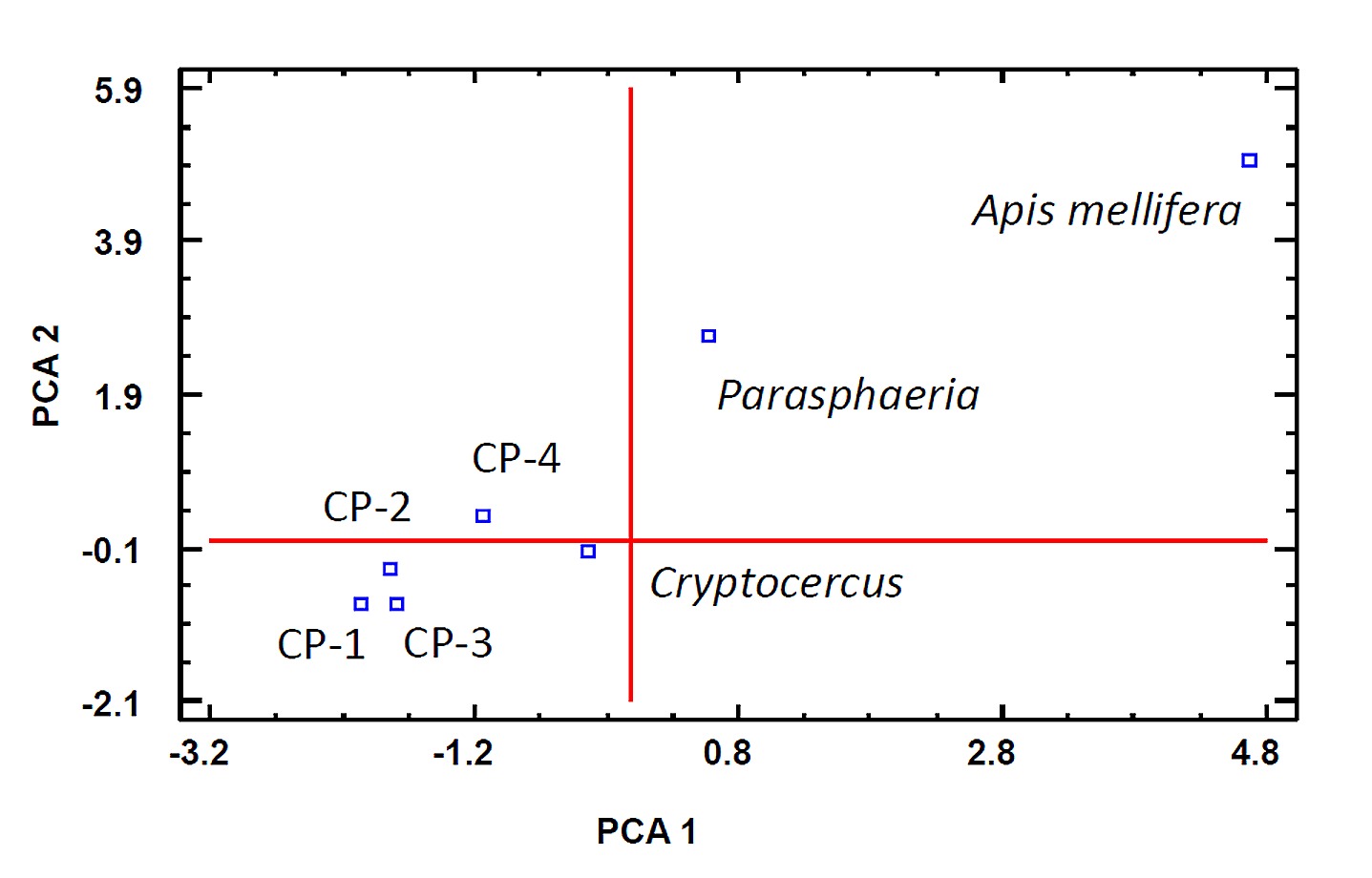
Relative abundance of class-phyla data were analyzed using the statistical program R v3.1.3. Kolmogorov-Smirnov test, a nonparametric test, were used to determine statistically significant difference between two samples based on a confidence level of 95.0% (P < 0.05 was considered statically significant). A principal components analysis (PCA) was performed to describe the relative abundance at Family level for the Cryptocercus (this work plus other Cryptocercus metagenomes published in the data base; see results) and Parasphaeriacockroaches. PCA data were treated with the pairwise and standardized options. Two components were extracted, and they accounted for 78.86% of the variability in the data.
Results
Gut bacterial community in Cryptocercus and Parasphaeria
After quality control filtering (see Material and Methods), 3519 and 2744 pyrosequencing reads were obtained from the Cryptocercus and Parasphaeria hindgut, respectively. Operational taxonomic units (OTUs) were defined for multiple cutoffs up to the distance threshold (0.03, 0.05, and 0.1). Rarefaction curves allowed the calculation of OTU richness for both the Cryptocercus and the Parasphaeria hindgut.
The calculated rarefaction curves estimated for the two cockroaches showed that the sampling reached an asymptote at the 0.10% genetic distance level (approximately at the family level), indicating that a reasonable number of OTUs had been acquired and that more intensive sampling would likely yield only a few additional OTUs [32,33]. However, rarefaction analysis at either the genus (0.05 distance threshold) or the species (0.03) level indicated that the number of reads analyzed was not sufficient to describe bacterial diversity within the cockroach gut. A total of 1150 and 889 SSU reference OTUs for Cryptocercus and Parasphaeria respectively were obtained. The representative reads (the longest read of each OUT defined at 97% sequence similarity) are compared to the SILVA reference datasets of the small- (16S/18S) subunit rDNA. The most abundant bacterial phyla in both cockroaches were Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria. The phylum Firmicutes was dominated by members of the Clostridia class, and Bacteroidetes mostly by members of the class Bacteroidia. The class profiles of Proteobacteria differed between Cryptocercus and Parasphaeria. In Cryptocercus, Alphaproteobacteria and Betaproteobacteria dominated whereas in Parasphaeria, members of the Alphaproteobacteria were the most abundant, followed by Deltaproteobacteria, and Gammaproteobacteria (Fig 1). The phylum candidatusSaccharibacteria (formerly known as Candidatus Division TM7) were present in Cryptocercus(4% of total bacterial population) but not in Parasphaeria. In Cryptocercus and Parasphaeria, spirochetes represented 1–2% of the total bacterial population. Elusimicrobia phyla (formerly Termite Group 1) represented less than 1% of the total bacterial population in Cryptocercus but they were not detected in Parasphaeria. On the other hand, Parasphaeria contained Deferribacteres and Fibrobacteres that represented less than 0.5% relative abundance Bacteria. Relative abundance class-taxon from Crytocercus was compared with relative abundance class-taxon from other Cryptocercus metagenomes deposited on the NCBI base data. A total of 5 different populations of Cryptocercus were analyzed (Cryptocercus this work; CP-1, CP-2, CP-3 from BioProject accession number PRJNA238270 [17]; and CP-4 from BioProject PRJNA217467) [34] (Fig 1). Relative abundance and population distribution were different among Cryptocercus indicating the “individual” variation of their microbiota, but Kolmogorov-Smirnov test showed that there were not significant differences at 95% confidence level (P ≥ 0.05) between Cryptocercus and CP-1; Cryptocercus and CP-2; Cryptocercus and CP-3 and Cryptocercus and CP-4. But there were significant differences (P < 0.05) between Cryptocercus and Parasphaeria (similar result was obtained using the unweighted UniFrac analysis, see below). PCA analysis revealed similarities among the bacterial microbiota of the different Cryptocercus metagenomes analyzed (S1 Fig). In this case, we considered “individual cockroach” as an individual member that probably represented the gut microbiota from a “like-colony” (several individuals living together), because cockroaches can be considered a gregarious insect [35,36].
Fig 1. Heatmap of relative abundance of phyla-class bacterial composition of the hindguts of the xylophagous cockroaches Cryptocercus and Parasphaeria.
Cryptocercus cockroaches: Cryptocercus (this work), CP-1, CP-2, CP-3 (Bioproject PRJNA238270) and CP-4 (PRJNA217467); and Parasphaeria (this work).
At 0.03 distances, Shannon’s diversity index showed that the intestinal tracts of Cryptocercusand Parasphaeria, 5.6 and 5.89 respectively, support a higher diversity community of bacteria similar to other wood or herbivorous-feeding insects [34,37] Bacterial community similarities between Cryptocercus and Parasphaeria were quantified based on phylogenetic relatedness by unweighted UniFrac. The analyses indicated that the composition of the bacterial communities from the two cockroaches differed significantly (P < 0.03). But, the Venn diagram generated by Mothur v1.36.1 from Cryptocercus and Parasphaeria at a genetic distance of 0.10 showed that six OTUs were shared and were related phylogenetically one to the phyla Spirochaetes (related to Treponema cluster I), two Bacteroidetes (Family Porphyromonadaceae belonging to Dysgonomonas and Parabacteroides genera) and tree Firmicutes (class Clostridia, uncultured bacteria of the Family XIII) (Fig 2). S1 and S2 Tables indicated the different OTUs and their taxonomic identification at genus level from Cryptocercus and Parasphaeria metagenomes, respectively.
Fig 2. Venn diagram.
Parasphaeria and Cryptocercus shared 6 OUT at 0.10 distances. Figure showed the two cockroaches. (Photo by M. Berlanga and R. Duro).
OTUs assigned to Spirochaetes
Spirochetal OTUs from Cryptocercus and Parasphaeria fell into three clusters, designated Treponema-termite clusters I, II, and III (Fig 3). Treponema-termite cluster I comprises both ectosymbionts attached to protists and free-swimming gut spirochetes from lower and higher termites. OTUs from Cryptocercus and Parasphaeria were grouped with free-swimming Treponema based on their affiliation with sequences of the isolates Treponema primitia or T. azotonutricium and other sequences of Treponema from higher termites (Fig 3). Sequences obtained from other metagenomes of Cryptocercus (CP1; CP2; CP3) Bioproject PRJNA238270 also cluster with pyrotags detected in this work. The second cluster, Treponema-termite cluster II, is much smaller than Treponema-termite cluster I and also much less diverse. It contains only sequences from lower termites and generally they were described in Reticulitermes and Hodotermopsis termites [38,39]. One OTU from Parasphaeria grouped with several sequences previously reported as belonging to Treponema-termite cluster II. Several OTUs from Cryptocercus and Parasphaeria grouped with Treponema-termite cluster III that contained Treponema sequences from other cockroaches and from higher termites (Fig 2). Data suggested that Spirochaetes from Cryptocercus and Parasphaeria could be free living bacteria present in the cockroaches before acquisition of flagellates’ protists by Cryptocercuscockroaches. Treponema detected in Cryptocercus and Parasphaeria were different to other Treponema described in other habitats such as human oral cavity (Fig 3).
Fig 3. Phylogenetic tree based on maximum-parsimony (MP) and maximum-likelihood (ML) analyses depicting the relationship among the pyrotags affiliated with the Treponema I, II, and III clades in termites and cockroaches.
Reference OTU from Cryptocercus and Parasphaeria were in bold. Other references OTU were obtained from Cryptocercus CP-1, CP-2 and CP-3 (Bioproject PRJNA238270) and Reticulitermes grassei [68]. In parentheses, the number of OTUs found repeatedly (at 0.05% genetic distance level). One thousand bootstrap trees were generated; bootstrap confidence levels, as percentages (only values >50%), are shown at tree nodes.
OTUs assigned to Bacteroidetes
In Cryptocercus and Parasphaeria, the dominant class was Bacteroidia and the family Porphyromonadaceae. OTUs from Cryptocercus were related to Bacteroidetes previously described sequences from different species of the protists Barbulanympha, Urinympha, Hoplonympha, and Mixotricha. Both Barbulanympha and Urinympha occur exclusively in the gut of Cryptocercus [40]. Several OTUs reference sequences from different cockroaches and termites were closely related to the before mentioned cluster. OTUs from Crytocercus also clustered with ectosymbiont Candidatus Symbiotrix of the protist Dinenympha. No representatives OTUs from Cryptocercus or Parasphaeria were clustered with other sequences belonging to termite symbionts protists Pseudotrichonympha or Streblomastix (Fig 4). Candidatus Symbiothrix, Dysgonomonas, Parabacteroides, Paludibacter and Tannerella were the major genera detected based on the Silva database. Porphyromonadaceae detected in cockroaches and termites were phylogenetically different from others obtained from cattle intestinal tracts (Fig 4).
Fig 4. Phylogenetic tree based on maximum-parsimony (MP) and maximum-likelihood (ML) analyses depicting the relationship among the pyrotags affiliated with Bacteroidetes.
Reference OTU from Cryptocercus and Parasphaeria were in bold. Other references OTU were obtained from Cryptocercus CP-1, CP-2 and CP-3 (Bioproject PRJNA238270), Reticulitermes grassei [68], Blattella germanica (Bioproject PRJEB3414) and Pyrrhcoris apterus (firebug) Bioproject PRJNA171139. In parentheses, the number of OTUs found repeatedly (at 0.10% genetic distance level). One thousand bootstrap trees were generated; bootstrap confidence levels, as percentages (only values >50%), are shown at tree nodes.
Clones assigned to Deltaproteobacteria
The phylum Proteobacteria represented 17.3 and 19.5 of relative pyrotags sequences obtained in Cryptocercus and Parasphaeria, respectively. Deltaproteobacteria represented 1.7 and 5.8 of the Proteobacteria sequences in Cryptocercus and Parasphaeria, respectively. The detected Deltaproteobacteria OTUs clustered with the families Desulfobacteraceae and Desulfovibrionaceae families. Desulfobacteraceae could be grouped in two clusters: one, grouped sequences obtained from cockroaches (Cryptocercus, Parasphaeria and Blattella germanica) and Reticulitermes termite; and second, OTUs detected from Cryptocercus and Parasphaeria clustered with syntrophic Deltaproteobacteria, such as Syntrophobacter spp. (Fig 5) OTUs from Cryptocercus belonging to the family Desulfovibrionaceace clustered with other sequences described from the termite gut related with the protist Trichonympha. In the Desulfovibrionaceae family, other group that contains sequences from several cockroaches and termites but were not related to symbionts of protists could be observed (Fig 5).
Fig 5. Phylogenetic tree based on maximum-parsimony (MP) and maximum-likelihood (ML) analyses depicting the relationship among the pyrotags affiliated with Deltaproteobacteria.
Reference OTU from Cryptocercus and Parasphaeria were in bold. Other references OTU were obtained from Cryptocercus CP-2 (Bioproject PRJNA238270), Reticulitermes grassei [68], Blattella germanica (Bioproject PRJEB3414) and Pyrrhcoris apterus(firebug) (Bioproject PRJNA171139). In parentheses, the number of OTUs found repeatedly (at 0.05% genetic distance level). One thousand bootstrap trees were generated; bootstrap confidence levels, as percentages (only values >50%), are shown at tree nodes.
Discussion
Insects contribute to an enormous diversity of symbiotic relationships. Microbial symbiosis probably played a central role in the evolutionary success of these organisms, allowing their adaptation to ecological niches that are nutritionally deprived and/or unbalanced (e.g., wood, plant sap or blood). The hindgut of insects is persistently colonized by opportunistic and commensal microbiota largely structured by exogenous (diet and local environment) and endogenous (gut environment) factors [17,21,41]. Transient bacteria acquired from the food and environment sources may complicate the apparent composition of gut microbial communities, but dynamic core gut microbiota (commensal) have been maintained even after environmental shifts [42–45]. Some termite gut-specific bacterial lineages have been observed that were not detected on environmental soil [45]. In this work we studied specific bacterial groups (phyla such as Spirochaetes, Bacteroidetes and Deltaproteobacteria) that have been described as a characteristic microbiota in the gut of termites and Cryptocercus [34,41].
Comparison of the relative abundance of class taxa among different Cryptocercusmetagenomes (our work and four Cryptocercus from the Bioprojects PRJNA238270 [17] and PRJNA217467 [37]) indicated individual variation of their microbiota, but there were no significant differences at 95% confidence level (P ≥ 0.05, Kolmogorov-Smirnov test) (Fig 1). “Individual” microbiota referred to several representatives’ members of a gregarious community of cockroaches living in a particular place, as it has been pointed out that there are few solitary cockroaches [35]. The “social” structure of Cryptocercus is the equivalent of a newly founded termite colony. After the eggs have hatched, adults feed the first few instars on hindgut fluids (proctodeal trophallaxis) [35]. The neonatal digestive tract is free of microbes, and the establishment of the full complement of microbial symbionts is a sequential process that varies in length between species. Typically, it is not complete until the third instar, when nutritional independence is possible, although close contact with adults is maintained [46]. Aggregation of the German cockroach, Blattella germanica, is regulated by fecal aggregation agents (pheromones), including volatile carboxylic acids [36]. The termite worker caste transfers food stomodeally (by regurgitation) and/or proctodeally (by excretion with the hindgut contents). Both oral trophallaxis (feeding) and coprophagy can promote a secure transmission of commensal microbiota between gregarious cockroaches [47–50] or members of a colony (termites).
In Dictyoptera, the transition from an omnivorous to a wood-feeding lifestyle had a strong impact on the microbial community structure, as observed in Cryptocercus and (lower) termites which included the acquisition of cellulolytic flagellates [17,41]. The complete loss of all flagellates in higher termites constituted another hallmark in the evolution of Isoptera [34,41]. But, other wood-feeding cockroaches, such as Panesthia spp., Salganea spp. and Parasphaeria spp. did not support the characteristic community of gut protists observed in the cockroach Cryptocercus [51,52]. In omnivorous cockroaches such as Periplaneta americana, their cellulose-rich diets have favored high protist numbers (e.g., Nyctotherus ovalis, Ciliophara), resulting in high cellulase activity. In fact, N. ovalis is responsible for most of the cellulolytic activity of P. americana [53]. The hindguts of the wood-feeding cockroach subfamily Panesthiinae harbor ciliated protists but they are probably not associated with the digestion of wood; rather, they are close related to the genus Nyctotherus, present in other cockroaches [54,55]. In thegenus Parasphaeria, while lacking the specific gut flagellates harbored in lower termites and Cryptocercus, many bacterial taxa were found. Of particular interest are Spirochaetes (Fig 3), Bacteroidetes (Fig 4), and Deltaproteobacteria (Fig 5), which are closely related to bacterial symbionts that specifically colonize the surface or interior of termite gut flagellates [6,56,57]. The sequences of Spirochaetes, Bacteroidetes and Deltaproteobacteriadetected in Parasphaeria and other cockroaches likely represent free-living relatives present in a common ancestor of cockroaches before their association with specific protists [44,57,58].
In Cryptocercus and Parasphaeria, Spirochaetes accounted for 1–2% of the total population. They are rare or not detected in omnivorous cockroaches, such as Shelfordella, Periplaneta, or in the xylophagous Panesthia angustipennis [37,44,54,59,60], but they are abundant in termites, especially in higher termites [61]. Spirochetes specialize in metabolic interactions with their hosts or other co-occurring microorganisms [62]. The main compounds produced by spirochetes are acetate, H2, and CO2, all of which are consumed by sulfate-reducing bacteria and methanogens (with both groups represented in termites). Acetate produced by the gut microbiota supports up to 100% of the respiratory requirement of termites [63]. Spirochetes from termite hindguts possess homologues of a nitrogenase gene (nifH) and exhibit nitrogenase activity [57]. This observation implicates spirochetes in the nitrogen nutrition of termites, whose food is typically low in nitrogen. Spirochete populations can stably maintain the gut habitat by supplying carbon sources and electron donors to other resident microbial populations and to the host.
Bacteroidetes OTUs from Cryptocercus grouped closely with phylotypes previously described from different species of the protist Barbulanympha. The genera Candidatus Symbiothrix, Dysgonomonas, Parabacteroides, Paludibacter and Tannerella were the major genera identified. In Parasphaeria, based on the Silva database, the major genera were Paludibacter, Parabacteroides and Candidatus Symbiothrix. These genera may represent the termite-specific bacterial lineages reported in termites [64,65]. Members of Bacteroidetes are thought to be specialized in the degradation of complex organic matter, including lignocellulosic compounds [66]. Bacteroidetes were also related to diazotrophic bacteria such as Azobacteroides pseudotrichonympha that provide amino acids and cofactors for the nutrition of the host protist and of the cockroach host [51,57].
Deltaproteobacteria OTUs in Cryptocercus and Parasphaeria hindguts were assigned to the families Desulfobacteriaceae and Desulfovibrionaceae. Both groups are strict anaerobes that are capable of sulfate-reduction. Sulfate-reducing bacteria (SRB) are crucial to the final step of carbon recycling and to the sulfur cycle in anaerobic ecosystems [67]. In addition to being important hydrogenotrophs (H2-consuming microorganisms), SRB contribute to the anoxic milieu of the gut by producing hydrogen sulfide and by removing oxygen together with hydrogen or low molecular weight organic or reduced sulfur compounds. H2 fluxes almost certainly play a significant role in shaping community structure, as H2 is both widely utilized as a microbial substrate and strongly influences the thermodynamics of the reactions in which it participates [67].
The molecular characterization carried out in this work revealed that bacterial community differ significantly between Cryptocercus and Parasphaeria; but they shared several bacterial genera found in other termites and cockroaches. Of special interest were several common OTUs detected in the intestinal tract belonging to Spirochaetes, Bacteroidetes and Firmicutes(Clostridia) that could represent a core microbiota essential to hydrolyze plant compounds and to provide nitrogen sources such as amino acids to their host (Fig 2). Wood-feeding Cryptocercus and Parasphaeria were phylogenetically distant cockroaches, but several bacterial groups were present in both cockroaches and were shared also with termites. Those bacteria may derive from the microbiota of a common ancestor before the diversification of cockroaches, subsequently diversified and adapted in each host. The microbiota depends on the hosts’ feeding behavior and secretions. The composition and physical form of the food changes as it passes down the gastrointestinal tract, offering microbes at different locations a changing complement of nutrients. Finally, the host obtains multiple nutrients in appropriate quantities and balance to optimally perform their biological function.
Supporting Information
S1 Fig. Principal Component Analysis (PCA) for Cryptocercus and Parasphaeriacockroaches based on relative abundance of bacterial taxa determined with Silva base data.
Cryptocercus cockroaches: Cryptocercus (this work), CP-1, CP-2, CP-3 (Bioproject PRJNA238270) and CP-4 (PRJNA217467); Parasphaeria (this work); Apis mellifera(PRJNA82239) as an out group of the Dictyoptera insect.
doi:10.1371/journal.pone.0152400.s001
(TIF)
S1 Table. Representative OTUs at 0.05 distance for Cryptocercus.
doi:10.1371/journal.pone.0152400.s002
(PDF)
S2 Table. Representative OTUs at 0.05 distance for Parasphaeria.
doi:10.1371/journal.pone.0152400.s003
(PDF)
Author Contributions
Conceived and designed the experiments: MB. Performed the experiments: MB JC. Analyzed the data: MB RG. Wrote the paper: MB RG. Participated in processing and quality control of the pyrotags obtained from 454-pyrosequencing: CL.
References
- 1.Moya A, Peretó J, Gil R, Latorre A. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet. 2008; 9:218–229. doi: 10.1038/nrg2319. pmid:18268509
- 2.Engel P, Moran NA. The gut microbiota of insects–diversity in structure and function. FEMS Microbiol Rev. 2013; 37: 699–735. doi: 10.1111/1574-6976. pmid:23692388
- 3.Jones RT, Sanchez LG, Fierer N. A cross-taxon analysis of insect-associated bacterial diversity. PLoS one. 2013; 8(4):e612118. doi: 10.1371/journal.pone.0061218.
- 4.Inward D, Beccaloni G, Eggleton P. Death of an order: a comprehensive molecular phylogenetic study confirms that termites are eusocial cockroaches. Biol. Lett. 2007; 3:331–335. doi: 10.1098/rsbl.2007.0102. pmid:17412673
- 5.Trautwein MD, Wiegmann BM, Beutel R, Kjer KM, Yeates DK. Advances in insect phylogeny at the dawn of the postgenomic era. Annu Rev Entomol. 2012; 57: 449–468. doi: 10.1146/annurev-ento-120710-100538. pmid:22149269
- 6.Ohkuma M. Symbioses of flagellates and prokaryotes in the gut of lower termites. Trends Microbiol. 2008; 16:345–352. doi: 10.1016/j.tim.2008.04.004. pmid:18513972
- 7.Berlanga M, Paster BJ, Guerrero R. The taxophysiological paradox: changes in the intestinal microbiota of the xylophagous cockroach Cryptocercus punctulatusdepending on the physiological state of the host. Int Microbiol. 2009; 12:227–236. doi: 10.2436/20.1501.01.102. pmid:20112227
- 8.Guerrero R, Margulis L, Berlanga M. Symbiogenesis: the holobiont as a unit of evolution. Int Microbiol. 2013; 16:133–143. doi: 10.2436/20.1501.01.188. pmid:24568029
- 9.Klass K-D, Nalepa C, Lo N. Wood-feeding cockroaches as models for termite evolution (Insecta: Dictyoptera): Cryptocercus vs. Parasphaeria boleiriana. Mol Phylolgenet Evol. 2008; 46:809–817. doi: 10.1016/j.ympev.2007.11.028
- 10.Lo N, Bandi C, Watanabe H, Nalepa C, Beninati T. Evidence for cocladogenesis between diverse dictyopteran lineages and their intracellular endosymbionts. Mol Biol Evol. 2003; 20:907–913. pmid:12716997 doi: 10.1093/molbev/msg097
- 11.Pellens R, Grandcolas P, Da Silva-Neto ID. A new and independently evolved case of xylophagy and the presence of intestinal flagellates in the cockroach Parasphaeria boleiriana (Dictyoptera, Blaberidae, Zetoborinae) from the remnants of the Brazilian. Can J Zool. 2002; 80:350–359. doi: 10.1139/z01-230
- 12.Kambhampati S, Peterson AT. Ecological niche conservation and differentiation in the wood-feeding cockroaches, Cryptocercus, in the United States. Biol J Linnean Soc. 2007; 90:457–466. doi: 10.1111/j.1095-8312.2006.00734.x
- 13.Grandcolas P, Pellens R. A new species of the cockroach genus Parasphaeria(Dictyoptera: Blattaria: Blaberidae) from the Atlantic forest in Brazil. Trans Am Entomol Soc. 2002; 128:23–29.
- 14.Pellens R, D’Haese CA, Bellés X, Piulachs M-D, Legendre F, Wheeler WC, et al. The evolutionary transition from subsocial to eusocial behaviour in Dictyoptera: phylogenetic evidence for modification of the “shift-in-dependent-care” hypothesis with a new subsocial cockroach. Mol Phyl Evol. 2007; 43:616–626. doi: 10.1016/j.ympev.2006.12.017
- 15.Hongoh Y. Toward the functional analysis of uncultivable, symbiotic microorganisms in the termite gut. Cell Mol Life Sci. 2011; 68:1311–1325. doi: 10.1007/s00018-011-0648-z. pmid:21365277
- 16.Rosengaus EB, Zecher CN, Schultheis KF, Brucker RM, Bordenstein SR. Disruption of the termite gut microbiota and its prolonged consequences for fitness. Appl Environ Microbiol. 2011; 77:4303–4312. doi: 10.1128/AEM.01886-10. pmid:21571887
- 17.Tai V, James ER, Nalepa CA, Scheffrahn RH, Perlman SJ, Keeling PJ. The role of host phylogeny varies in shaping microbial diversity in the hindguts of lower termites. Appl Environ Microbiol. 2015; 81:1059–1070. doi: 10.1128/AEM.02945-14. pmid:25452280
- 18.Berlanga M, Paster BJ, Guerrero R. Coevolution of symbiotic spirochete diversity in lower termites. Int Microbiol. 2007; 10:133–239. doi: 10.2436/20.1501.01.19. pmid:17661292
- 19.Minard G, Tran F-H, Dubost A, Tran-Van V, Mavingui P, Moro CV. Pyrosequencing 16S rRNA genes of bacteria associated with wild tiger mosquito Aedes albopictus: a pilot study. Front Cell Infect Microbiol. 2014; 4:59. doi: 10.3389/fcimb.2014.00059. pmid:24860790
- 20.Middelbos IS, Boler BMV, Qu A, White BA, Swanson KS, Fahey GC. Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS ONE. 2010; 5(3):e9768. doi: 10.1371/journal.pone.0009768. pmid:20339542
- 21.Yun JH, Roh SW, Whon TW, Jung MJ, Kim MS, Park DS, et al. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl Environ Microbiol. 2014; 80:5254–5264. doi: 10.1128/AEM.01226-14. pmid:24928884
- 22.Futami R, Muñoz-Pomer L, Viu JM, Dominguez-Escriba L, Covelli L, Bernet GP, et al. GPRO The professional tool for annotation, management and functional analysis of omic databases. Biotechvana Bioinformatics 2011-SOFT3. 2011.
- 23.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011; 17:10–12. doi: 10.14806/ej.17.1.200.
- 24.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011; 27:863–864. doi: 10.1093/bioinformatics/btr026. pmid:21278185
- 25.FastQC. 2012; http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- 26.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009; 75:7537–7541. doi: 10.1128/AEM.01541-09. pmid:19801464
- 27.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009; 25:1335–1337. doi: 10.1093/bioinformatics/btp157. pmid:19307242
- 28.Nicholas KB, Nicholas HB Jr, Deerfield DW. GeneDoc: Analysis and visualization of genetic variation. EMBNEW. NEWS. 1997; 4:14. [http://www.psc.edu/biomed/genedoc].
- 29.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012; 28:3150–3152. doi: 10.1093/bioinformatics/bts565. pmid:23060610
- 30.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013; 41:D590–596. doi: 10.1093/nar/gks1219. pmid:23193283
- 31.Lozupone C, Hamady M, Knight R. UniFrac–an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006; 7:371. pmid:16893466
- 32.Yarza P, Ludwig W, Euzéby J, Amann R, Schleifer K- H, Glöckner FO, et al. Update of the all-species living tree project based on 16S and 23S rRNA sequence analyses. Syst Appl Microbiol. 2010; 33:291–299. doi: 10.1016/j.syapm.2010.08.001. pmid:20817437
- 33.Sun Y, Cai Y, Liu L, Yu F, Farrell ML, Mckendree W, et al. ESPRIT: estimating species richness using large collections of 16S rRNA pyrosequences. Nucleic Acids Res. 2009; 37:e76. doi: 10.1093/nar/gkp285. pmid:19417062
- 34.Dietrich C, Köhler T, Brune A. The cockroach origin of the termite gut microbiota: patterns in bacterial community structure reflect major evolutionary events. Appl Environ Microbiol. 2014; 80:2261–2269. doi: 10.1128/AEM.04206-13. pmid:24487532
- 35.Nalepa CA. Origin of termite eusociality: trophallaxis integrates the social, nutricional, and microbial environments. Ecol Entomol. 2015; 40:323–335. doi: 10.1111/een.12197.
- 36.Wada-Katsumata A, Zurek L, Nalyanya G, Roelofs WL, Zhang A, Schal C. Gut bacteria mediate aggregation in the German cockroach. Proc Natl Acad Sci USA. 2015; 112:15578–15683. doi: 10.1073/pnas.1504031112.
- 37.Scully ED, Geib SM, Carlson JE, Tien M, McKenna D, Hoover K. Functional genomics and microbiome profiling of the Asian longhorned beetle (Anoplophora glabripennis) reveal insights into the digestive physiology and nutritional ecology of wood feeding beetles. BMC Genomics. 2014; 15:1096. doi: 10.1186/1471-2164-15-1096. pmid:25495900
- 38.Lilburn TG, Schmidt TM, Breznak JA. Phylogenetic diversity of termite gut spirochaetes. Environ Microbiol. 1999; 1:331–345. pmid:11207751 doi: 10.1046/j.1462-2920.1999.00043.x
- 39.Iida T, Ohkuma M, Ohtoko K, Kudo T. Symbiotic spirochetes in the termite hindgut: phylogenetic identification of ectosymbiotic spirochetes of oxymonad protists. FEMS Microbiol Ecol. 2000; 34:17–26. pmid:11053732 doi: 10.1111/j.1574-6941.2000.tb00750.x
- 40.Noda S, Inoue T, Hongoh Y, Kawai M, Nalepa CA, Vongkaluang C, et al. Identification and characterization of ectosymbionts of distinct lineages in Bacteroidales attached to flagellated protists in the gut of termites and a wood-feeding cockroach. Environ Microbiol. 2006; 8:11–20. pmid:16343317 doi: 10.1111/j.1462-2920.2005.00860.x
- 41.Brune A, Dietrich C. The gut microbiota of termites: Digesting the diversity in the light of ecology and evolution. Annu Rev Microbiol. 2015; 69:145–166. doi: 10.1146/annurev-micro-092412-155715. pmid:26195303
- 42.Sudakaran S, Salem H, Kost C, Kaltenpoth M. Geographical and ecological stability of the symbiotic mid-gut microbiota in European firebugs, Pyrrhocoris apterus (Hemiptera, Pyrrhocoridae). Mol Ecol. 2012; 21:6134–6151. doi: 10.1111/mec.12027. pmid:23017151
- 43.Boucias DG, Cai Y, Sun Y, Lietze VU, Sen R, Raychoudhury R, et al. The hindgut lumen prokaryotic microbiota of the termite Reticulitermes flavipes and its responses to dietary lignocellulose composition. Mol Ecol. 2013; 22:1836–1853. doi: 10.1111/mec.12230. pmid:23379767
- 44.Schauer C, Thompson C, Brune A. Pyrotag sequencing of the gut microbiota of the cockroach Shelfordella lateralis reveals a highly dynamic core but only limited effects of diet on community structure. PLoS ONE. 2014; 9:e85861. doi: 10.1371/journal.pone.0085861. pmid:24454939
- 45.Makonde HM, Mwirichia R, Osiemo Z, Boga HI, Klenk H-P. 454 Pyrosequencing-based assessments of bacterial diversity and community structure in termite guts, mounds and surrounding soils. Springerplus. 2015; 4:471. doi: 10.1186/s40064-015-1262-6. pmid:26355944
- 46.Nalepa CA. Early development of nymphs and establishment of hindgut symbiosis in Cryptocercus punctulatus (Dictyoptera: Cryptocercidae). Ann Entomol Soc Amer. 1990; 83:786–789. doi: 10.1093/aesa/83.4.786
- 47.Berlanga M, Paster BJ, Guerrero R. Coevolution of symbiotic spirochete diversity in lower termites. Int Microbiol. 2007; 10:133–139. pmid:17661292
- 48.Brune A. Symbiotic digestion of lignocellulose in termites guts. Nat Rev Microbiol. 2014; 12:168–180. doi: 10.1038/nrmicro3182. pmid:24487819
- 49.Rahman NA, Parks DH, Willner DL, Engelbrektson AL, Goffredi SK, Warnecke F, et al. A molecular survey of Australian and North American termite genera indicates that vertical inheritance is the primary force shaping termite gut microbiomes. Microbiome. 2015; 3:5. doi: 10.1186/s40168-015-0067-8. pmid:25830022
- 50.Diouf M, Roy V, Mora P, Frechault S, Lefebvre T, Hervé V, et al. Profiling the succession of bacterial communities throughout the life stages of a higher termite Nasutitermes arborum (Termitidae, Nasutitermitinae) using 16S rRNA gene pyrosequencing. PLoS ONE. 2015; 10(10):e0140014 doi: 10.1371/journal.pone.0140014. pmid:26444989
- 51.Noda S, Hongoh Y, Sato T, Ohkuma M. Complex coevolutionary history of symbiotic Bacteroidades bacteria of various protist in the gut of termites. BMC Evol Biol. 2009; 9:158. doi: 10.1186/1471-2148-9-158. pmid:19586555
- 52.Ohkuma M, Noda S, Hongoh Y, Nalepa CA, Inoue T. Inheritance and diversification of symbiotic trichonymphid flagellates from a common ancestor of termites and the cockroach Cryptocercus. Proc R Soc B. 2009; 276:239–245. doi: 10.1098/rspb.2008.1094. pmid:18812290
- 53.Gijzen HJ, van der Drift C, Barugahare M, Huub JM, den Camp O. Effect of host diet and hindgut microbial composition on cellulolytic activity in the hindgut of the american cockroach, Periplaneta americana. Appl Environ Microbiol. 1994; 60:1822–1826. pmid:16349275
- 54.Bauer E, Lampert N, Mikaelyan A, Köhler Y, Maekawa K, Brune A. Physicochemical conditions, metabolites and community structure of the bacterial microbiota in the gut of Wood-feeding cockroaches (Blaberidae: Panesthiinae). FEMS Microbiol Ecol. 2015; 91:1–12. doi: 10.1093/femsec/fiu028.
- 55.Lynn DH, Wright ADG. Biodiversity and molecular phylogeny of Australian Clevelandella species (class Armophorea, order Clevelandellida, family Clevelandellidae), intestinal endosymbiotic ciliates in the wood-feeding roach Panesthia cribrata Saussure, 1864. J Eukaryot Microbiol. 2013; 60:335–341. doi: 10.1111/jeu.12037. pmid:23590673
- 56.Ohkuma M, Noda S, Hattori S, Lida T, Yuki M, Starns D, et al. Acetogenesis from H2plus CO2 and nitrogen fixation by an endosymbiotic spirochete of a termite-gut cellulolytic protist. Proc Natl Acad Sci USA. 2015; 112:10224–10230. doi: 10.1073/pnas.1423979112. pmid:25979941
- 57.Desai MD, Brune A. Bacteroidales ectosymbionts of gut flagellates shape the nitrogen-fixing community in dry-wood termites. ISME J. 2012; 6:1302–1313. doi: 10.1038/ismej.2011.194. pmid:22189498
- 58.Ikeda-Ohtsubo W, Faivre N, Brune A. Putatively free-living “Endomicrobia”—ancestors of the intracellular symbionts of termite gut flagellates? Environ Microbiol Rep. 2010; 2:554–559. doi: 10.1111/j.1758-2229.2009.00124.x. pmid:23766225
- 59.Bertino-Grimaldi D, Medeiros MN, Vieira RP, Cardoso AM, Turque AS, Silveira CB, et al. Bacterial community composition shifts in the gut of Periplaneta americana fed on different lignocellulosic materials. Springerplus. 2013; 2:609. doi: 10.1186/2193-1801-2-609. pmid:24324923
- 60.Carrasco P, Pérez-Cobas AE, van de Pol C, Baixeras J, Moya A, Latorre A. Succession of the gut microbiota in the cockroach Blattella germanica. Int Microbiol. 2014; 17:99–109. doi: 10.2436/20.1501.01.212. pmid:26418854
- 61.Köhler T, Dietrich C, Scheffrahn RH, Brune A. High-resolution analysis of gut environment and bacterial microbiota reveals functional compartmentation of the gut in wood-feeding higher termites (Nasutitermes spp.) Appl Environ Microbiol. 2012; 78:4691–4701. doi: 10.1128/AEM.00683-12. pmid:22544239
- 62.Rosenthal AZ, Matson EG, Eldar A, Leadbetter JR. RNA-seq reveals cooperative metabolic interactions between two termite-gut spirochete species in co-culture. ISME J. 2011; 5:1133–1142. doi: 10.1038/ismej.2011.3.
- 63.Leadbetter JR, Schmidt TM, Graber JR, Breznak JA. Acetogenesis from H2 plus CO2by spirochetes from termite guts. Science. 1999; 283:686–689. pmid:9924028 doi: 10.1126/science.283.5402.686
- 64.Shinzato N, Muramatsu M, Matsui T, Watanabe Y. Phylogenetic analysis of the gut bacterial microflora of the fungus-growing termite Odontotermes formosanus. Biosci Biotechnol Biochem. 2007; 71:906–915. pmid:17420599 doi: 10.1271/bbb.60540
- 65.Otani S, Mikaelyan A, Nobre T, Hansen LH, Kone N, Sørensen SJ, et al. Identifying the core microbial community in the gut of fungus-growing termites. Mol Ecol. 2014; 23:4631–4644. doi: 10.1111/mec.12874. pmid:25066007
- 66.Yuki M, Kuwahara H, Shintani M, Izawa K, Sato T, Starns D, et al. Dominant ectosymbiotic bacteria of cellulolytic protists in the termite gut also have the potential to digest lignocellulose. Environ Microbiol. 2015; 17:4942–4953. doi: 10.1111/1462-2920.12945. pmid:26079531
- 67.Dröge S, Limper U, Emtiazi F, Schönig I, Pavlus N, Drzyzga O, et al. In vitro and in vivo sulfate reduction in the gut contents of the termite Mastotermes darwiniensis and the rose-chafer Pachnoda marginata. J Gen Appl Microbiol. 2005; 51:57–64. doi: 10.2323/jgam.51.57. pmid:15942866
- 68.Berlanga M, Paster BJ, Grandcolas P, Guerrero R. Comparison of the gut microbiota from soldier and worker castes of the termite Reticulitermes grassei. Int Microbiol. 2011; 14:83–93. doi: 10.2436/20.1501.01.138. pmid:22069152
For further details log on website :
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0152400





No comments:
Post a Comment